Here are some instructions for making a basic installation of VASP 5.3.3 on Cray XE6. It applies specifically to the Cray XE6 at PDC called “Lindgren”, but Cray has a similar environment on all machines, so it might be helpful for other Cray sites as well.
First, download the prerequisite source tarballs from the VASP home page:
http://www.vasp.at/
You need both the regular VASP source code, and the supporting “vasp 5” library:
vasp.5.3.3.tar.gz
vasp.5.lib.tar.gz
I suggest to make a new directory called e.g. vasp.5.3.3, where you download and expand them. You would type commands approximately like this:
mkdir 5.3.3
cd 5.3.3
(download)
tar zxvf vasp.5.3.3.tar.gz
tar zxvf vasp.5.lib.tar.gz
Which compiler?
The traditional compiler for VASP is Intel’s Fortran compiler (“ifort”). Version 12.1.5 of ifort is now the “official” compiler, the one which the VASP developers use to compile the program. Unfortunately, ifort is one of the few compilers which can compile the VASP source unmodified, since the code contains non-standard Fortran constructs. To compile with e.g. gfortran, pgi, or pathscale ekopath, which theoretically could generate better code for AMD processors, source code modifications are necessary. So we will stick with Intel’s Fortran compiler in this guide. On the Cray machine, this module is called “PrgEnv-Intel”. Typically, PGI is the default preloaded compiler, so we have to swap compiler modules
module swap PrgEnv-pgi PrgEnv-intel
Check which version of the compiler you have by typing “ifort -v”:
$ ifort -v
ifort version 12.1.5
If you have the “PrgEnv-intel/4.0.46” module loaded, it should state “12.1.5”.
Which external libraries?
For VASP, we need BLAS, LAPACK, SCALAPACK and the FFTW library. On the Cray XE6, these are usually provided by Cray’s own “libsci” library. This library is supposedly specifically tuned for the Cray XE6 machine, and should offer good performance.
Check that the libsci module is loaded:
$ module list
...
xt-libsci/11.1.00
...
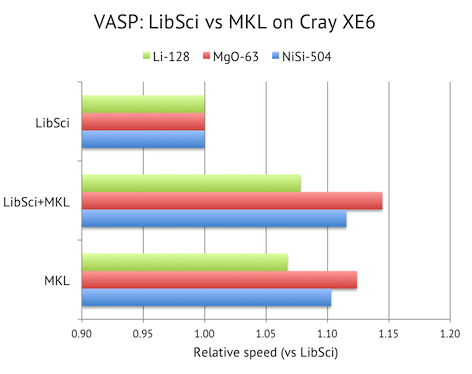
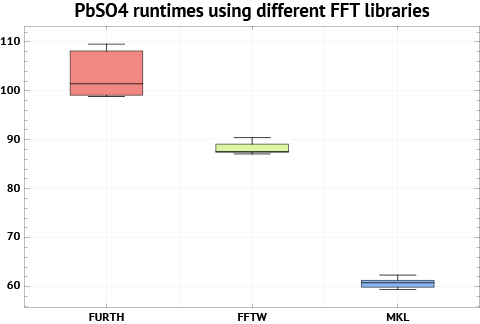
Normally, you combine libsci with the FFTW library. But I would recommend using the FFT routines from MKL instead, since they result in 10-15% faster overall speed in my benchmarks. Recent versions of MKL comes with FFTW3-compatible wrappers built-in (you don’t need to compile them separately), so by linking with MKL and libsci in the correct order, you get the best from both worlds.
VASP 5 lib
Compiling the VASP 5 library is straightforward. It contains some timing and IO routines, necessary for VASP, and LINPACK. My heavy edited makefile looks like this:
.SUFFIXES: .inc .f .F
#-----------------------------------------------------------------------
# Makefile for VASP 5 library on Cray XE6 Lindgren at PDC
#-----------------------------------------------------------------------
# C-preprocessor
CPP = gcc -E -P -C -DLONGCHAR $*.F >$*.f
FC= ftn
CFLAGS = -O
FFLAGS = -O1 -FI
FREE = -FR
DOBJ = preclib.o timing_.o derrf_.o dclock_.o diolib.o dlexlib.o drdatab.o
#-----------------------------------------------------------------------
# general rules
#-----------------------------------------------------------------------
libdmy.a: $(DOBJ) linpack_double.o
-rm libdmy.a
ar vq libdmy.a $(DOBJ)
linpack_double.o: linpack_double.f
$(FC) $(FFLAGS) $(NOFREE) -c linpack_double.f
.c.o:
$(CC) $(CFLAGS) -c $*.c
.F.o:
$(CPP)
$(FC) $(FFLAGS) $(FREE) $(INCS) -c $*.f
.F.f:
$(CPP)
.f.o:
$(FC) $(FFLAGS) $(FREE) $(INCS) -c $*.f
Note that the Fortran compiler is always called “ftn” on the Cray (regardless of the module loaded), and the addition of the “-DLONGCHAR” flag on the CPP line. It activates the longer input format for INCAR files, e.g. you can have MAGMOM lines with more than 256 characters. Now compile the library with the “make” command and check that you have the “libdmy.a” output file. Leave the file here, as the main VASP makefile will include it directly from here.
Editing the main VASP makefile
I suggest that you start from the Linux/Intel Fortran makefile:
cp makefile.linux_ifc_P4 makefile
It is important to realise that the makefile is split in two parts, and is intended to be used in an overriding fashion. If you don’t want to compile the serial version, you should enable the definitions of FC, CPP etc in the second half of the makefile to enable parallel compilation. These will then override the settings for the serial version.
Start by editing the Fortran compiler and its flags:
FC=ftn -I$(MKL_ROOT)/include/fftw
FFLAGS = -FR -lowercase -assume byterecl
On Lindgren, I don’t get MKL_ROOT set by default when I load the PrgEnv-intel module, so you might have to do it by yourself too:
MKL_ROOT=/pdc/vol/i-compilers/12.1.5/composer_xe_2011_sp1.11.339/mkl
The step above is site-specific, so you should check where the Intel compilers are actually installed. If you cannot find any documentation, try inspecting the PATH after loading the module.
Then, we change the optimisation flags:
OFLAG=-O2 -ip
Note that we leave out any SSE/architectural flags, since these are provided automatically by the “xtpe-mc12” module (make sure that it is loaded by checking the output of module list).
We do a similar trick for BLAS/LAPACK by providing empty definitions for them:
# BLAS/LAPACK should be linked automatically by libsci module
BLAS=
LAPACK=
We need to edit the LINK variable to include Intel’s MKL. I also like to add more verbose output of the linking process, to check that I am linking the correct library. One simple way to do this is to ask the linker to say from where it picks up the ZGEMM subroutine (it should be from Cray’s libsci, not MKL).
LINK = -mkl=sequential -Wl,-yzgemm_
Now, move further down in the makefile, to the MPI section, and edit the preprocessors flags:
CPP = $(CPP_) -DMPI -DHOST=\"PDC-REGULAR-B01\" -DIFC \
-DCACHE_SIZE=4000 -DPGF90 -Davoidalloc -DNGZhalf \
-DMPI_BLOCK=262144 -Duse_collective -DscaLAPACK \
-DRPROMU_DGEMV -DRACCMU_DGEMV -DnoSTOPCAR
CACHE_SIZE is only relevant for Furth FFTs, which we do not use. The HOST variable is written out in the top of the OUTCAR file. It can be anything which helps you identify this compilation of VASP. The MPI_BLOCK variable needs to be set higher for best performance on the Cray XE6 interconnect. And finally, “noSTOPCAR” will disable the ability to stop a calculation by using the STOPCAR file. We do this to improve file I/O against the global file systems. (Otherwise, each VASP process will have to check this file for every SCF iteration.)
We will get Cray’s SCALAPACK linked in via libsci, so we set an empty SCA variable:
# SCALAPACK is linked automatically by libsci module
SCA=
Then activate the parallelized version of the fast Fourier transforms with FFTW bindings:
FFT3D = fftmpiw.o fftmpi_map.o fftw3d.o fft3dlib.o
Note that we do not need to link to FFTW explicitly, since it is included in MKL.
Finally, we uncomment the last library section for completeness:
LIB = -L../vasp.5.lib -ldmy \
../vasp.5.lib/linpack_double.o \
$(SCA) $(LAPACK) $(BLAS)
The full makefile is provided here.
Compiling
VASP does not have a makefile that supports parallel compilation. So in order to compile we just do:
make
If you really want to speed it up, you can try something like:
make -j4; make -j4; make -j4; make -j4;
Run these commands repeatedly until all the compiler errors are cleared (or write a loop in the bash shell). Obviously, this approach only works if you have a makefile that you know works from the start. When finished, you should find a binary called “vasp”.
Running
The Cray compiler environment produces statically linked binaries by default, since this is the most convenient way to run on the Cray compute nodes, so we just have to put e.g. the following in the job script to run on 384 cores (16 compute nodes).
aprun -n 384 /path/to/vasp
I recommend setting NPAR equal to the number of compute nodes on the Cray XE6. As I have shown before, it is possible to run really big VASP simulations on the Cray with decent scaling over 1000-2000 cores. If you go beyond 32 compute nodes, it is worth trying to run on only half the number of cores per node. So on a machine with 24 cores per node, you would ask for 768 cores, but actually run like this:
aprun -n 384 -N 12 -S 3 /path/to/vasp